The genetic connections among Brassica species have been identified and verified through experimental crosses between diploid and/or tetraploid Brassica crops, as well as microscopic or karyotyping examination during the synapsis stage of meiosis in these hybrids (Cheng et al. 2014; Zhang et al. 2021a). For instance, F1 plants with a chromosome count of n = 29 were produced from a cross between B. rapa (AA, n=10) andB. napus (AACC, n=19). Under microscopic examination, only nine typical pairs of synaptic chromosomes were observed in the meiotic cells of the F1 plants during the synapsis stage, corresponding to the two sets of chromosomes contributed by the B. rapa genome. The remaining ten chromosomes of B. napus, which were unable to form normal pairs of synaptic chromosomes in the F1 generation, were influenced by the chromosomes from the A genome of B. rapa. This provides credence to the hypothesis that these ten chromosomes in B. napus are homologous to the corresponding ten chromosomes in B. rapa (Morinaga 1929; Cheng et al. 2014).
Furthermore, similar experiments involving crosses between B. oleracea and B. napus demonstrated that the other nine chromosomes of B. napus are homologous to those of B. oleracea. Collectively, these findings support the conclusion that B. napus is a tetraploid resulting from interspecific hybridisation between B. oleracea and B. rapa (Cheng et al. 2014; Rong et al. 2019).
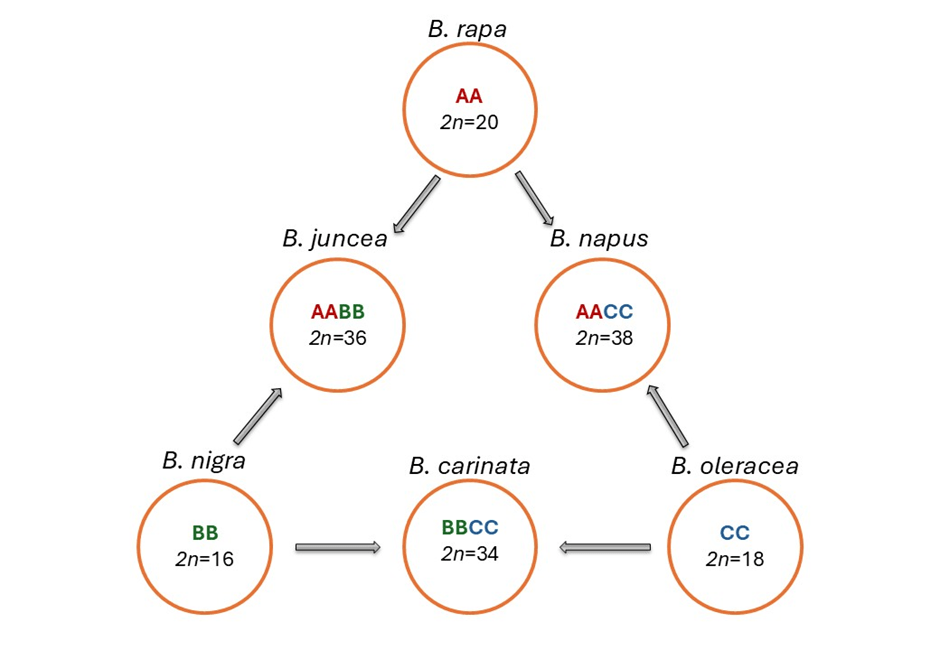
Figure 1: Genetic relationships among six Brassica species (U’s Triangle) (Rijal et al., 2026).
Based on experimental findings, the relationships among six Brassica species are succinctly described by the U’s Triangle model. In this model, the three diploid species, B. rapa, B. nigra, and B. oleracea, represent the fundamental A, B, and C genomes, respectively, positioned at the three vertices (Figure 1). The three allotetraploid species, B. juncea (AB), B. napus (AC), and B. carinata (BC), which resulted from hybridisation between these diploids, occupy the midpoints of the triangle’s edges. Thus, the U’s triangle model has been successfully applied to elucidate phylogenetic relationships within the Brassica species and has significantly contributed to their genetic studies (Cheng et al. 2014; Koh et al. 2017).
References:
Cheng F, Wu J, Wang X (2014) Genome triplication drove the diversification of Brassica plants. Hortic Res 1:14024. doi:10.1038/hortres.2014.24
Koh JC, Barbulescu DM, Norton S et al (2017) A multiplex PCR for rapid identification of Brassica species in the triangle of U. Plant Methods 13:1-8. doi:10.1186/s13007-017-0200-8
Morinaga T (1929) Interspecific hybridization in Brassica I. The cytology of F 1 hybrids of B. napella and various other species with 10 chromosomes. Cytologia 1:16-27. doi:10.1508/cytologia.1.16
Rijal, R., Sharm, A.K., Kumar, A. (2026). Economic Importance of Brassica Crops. In: Kumar, A., Koul, B. (eds) Crop Improvement Strategies in Brassica species: Applied Science. Springer, Singapore. https://doi.org/10.1007/978-981-95-3861-4_1
Rong H, Zhu S, Xie T et al (2019) Phenotypic and seed structural comparison in hybrids of different Brassica rapa and B. oleracea. Plant Breed 138:925-936. doi:10.1111/pbr.12748
Song X, Wei Y, Xiao D et al (2021) Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol186:388-406. doi:10.1093/plphys/kiab048
Zhang K, Mason AS, Farooq MA et al (2021a) Challenges and prospects for a potential allohexaploid Brassica crop. Theor Appl Genet 134:2711-2726. doi:10.1007/s00122-021-03845-8


Leave a comment